Mol2DSimi
Published:
Mol2DSimi
- Calculate molecular fingerprints (12 types) and similarity based on reference compounds
- Automated validation with several types of metrics: AUC, EF1%, EF5%, EF10%, F1 score, GH
- Statistical test for decision making: which type of fingerprint is suitable or reference compound that enhance performance
- Ensemble learning: stacking technique
Tanimoto similarity
\[T _{c}(A,B) = \frac{c}{a+b-c}\]- a: number of features present in molecule A
- b: number of features present in molecule B
- c: number of features shared by molecules A and B
Requirements
This module requires the following modules:
Installation
Clone this repository to use
Folder segmentation
Finally the folder structure should look like this:
Mol2DSimi (project root)
|__ README.md
|__ Mol2DSimi
|__ |__ Similarity.py
| |__ Simivalid.py
| |__ enrichment_factor.py
| |__ significantplot.py
|__ Image (saved images)
|__ Mol2DSimi.ipynb
|__ LICENSE
|......
Usage
import math
import numpy as np
import pandas as pd
from rdkit import Chem
import sys
sys.path.append('Mol2DSimi')
from Similarity import similarity_calculate
from enrichment_factor import Enrichment_Factor
from validation import similarity_validation
from tqdm import tqdm # progress bar
tqdm.pandas()
# 1. Similarity Calculation
simi = similarity_calculate(data = data, query= query, smile_col="CanonSmiles", active_col='Active')
simi.fit()
simi.plot()
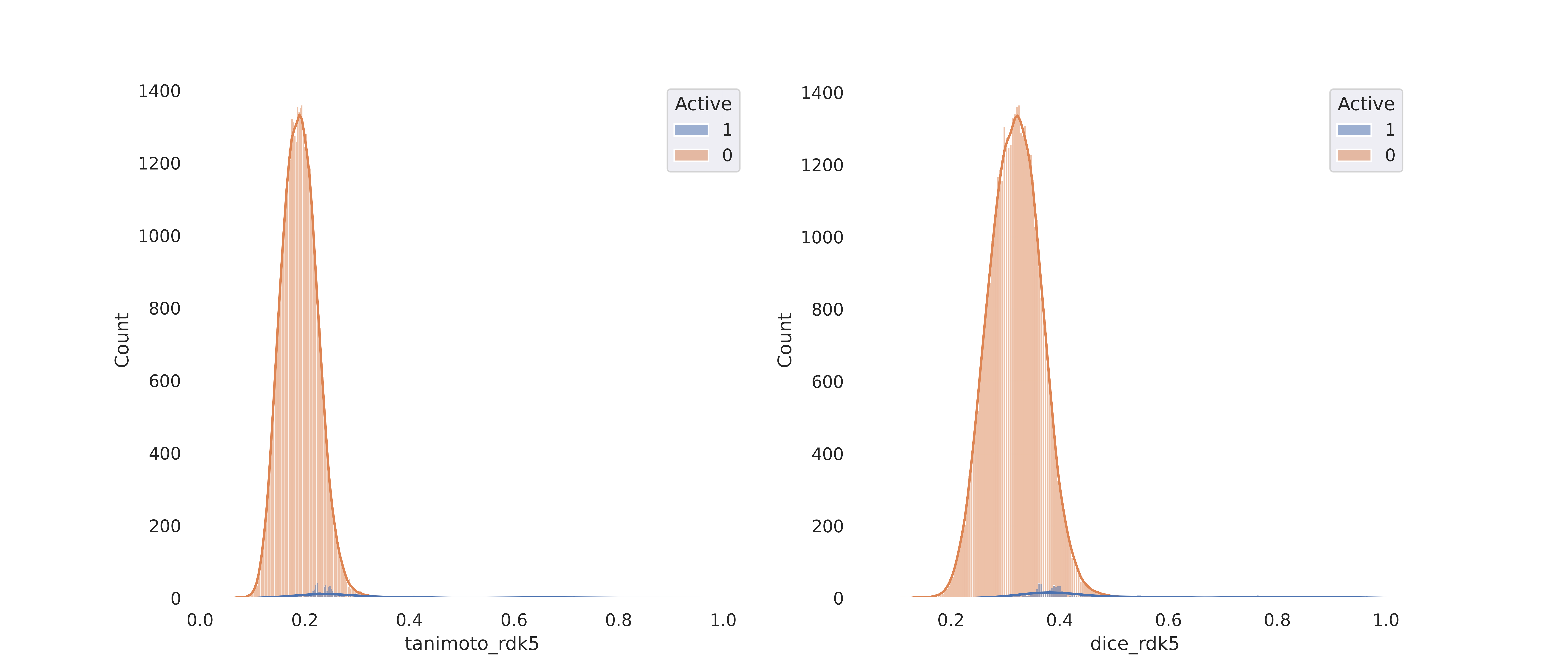
# 2. Valudation
valid = similarity_validation(data, active_col = 'Active', scores = 'tanimoto',plot_type = 'roc', figsize = (14,10), query =i )
valid.validation()
valid.visualize()
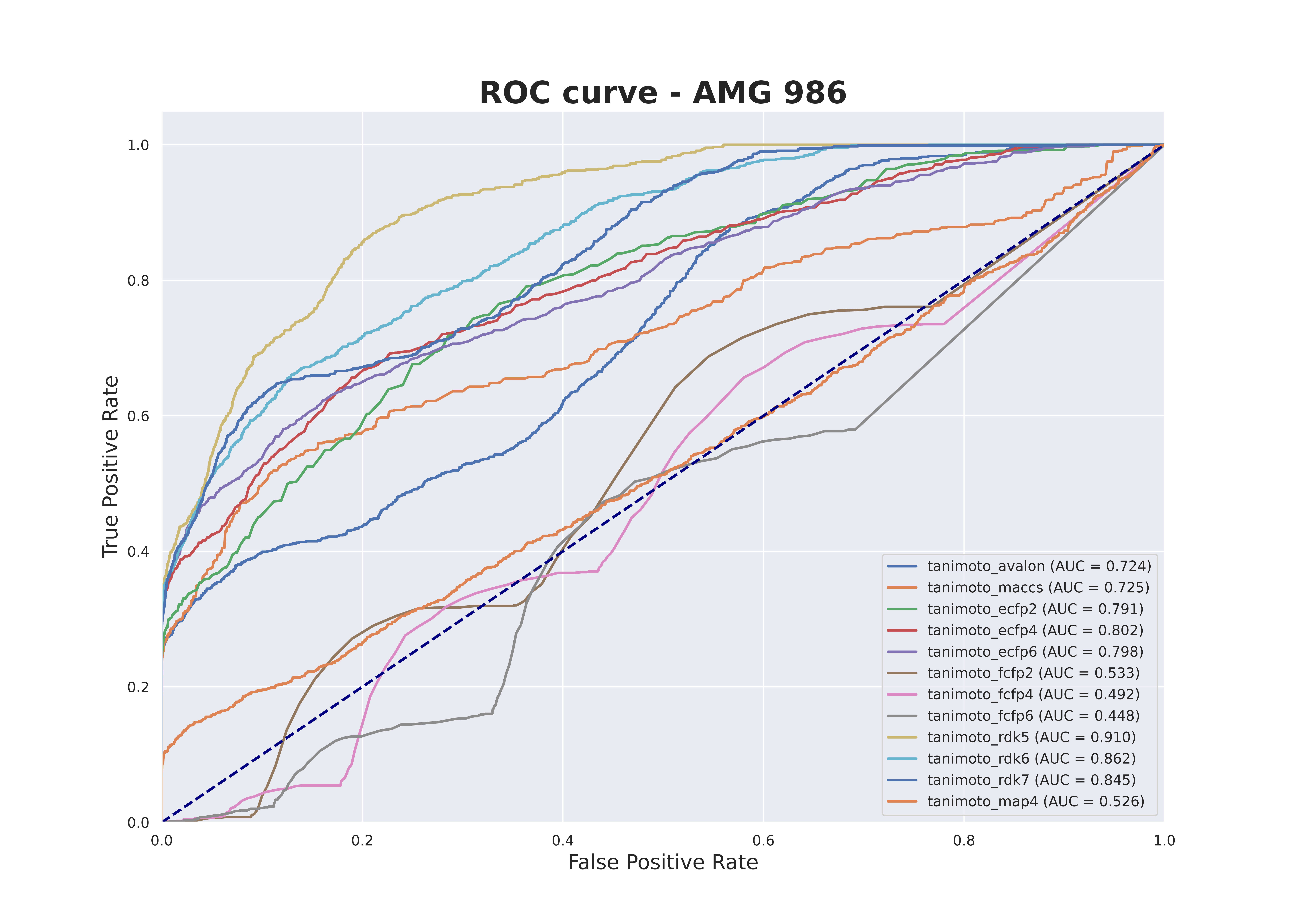
# 3. Compare fingerprints - automated pipeline
import warnings
warnings.filterwarnings('ignore')
from sklearn.model_selection import RepeatedStratifiedKFold, train_test_split
path = './Similarity/Data/Raw_data'
cv = RepeatedStratifiedKFold(n_repeats = 3, n_splits=10, random_state=42)
list_AUC = []
# query is a list of molecules format: CMF_019, AMG_986, BMS_986224
for i in query:
data = pd.read_csv(path+f'/{i.GetProp("_Name")}.csv')
data = data[col]
data_train, data_test = train_test_split(data, test_size=0.2, random_state=42, stratify = data.Active)
for train_index, test_index in cv.split(data_train.drop(['Active'], axis =1), data_train['Active']):
list_auc = []
model = []
test = data_train.iloc[test_index,:]
for i in col[1:]:
model.append(i)
fpr, tpr, _ = roc_curve(test['Active'], test[i])
roc_auc = round(auc(fpr, tpr),3)
list_auc.append(roc_auc)
list_AUC.append(list_auc)
AUC = pd.DataFrame(list_AUC, columns = model)
AMG_986 = AUC.iloc[30:60,:].reset_index(drop=True)
# post hoc
df_melt = pd.melt(AMG_986.reset_index(), id_vars=['index'], value_vars=AUC.columns)
df_melt.columns = ['index', 'Model', 'AUC']
pc =sp.posthoc_wilcoxon(df_melt, val_col='AUC', group_col='Model', p_adjust='holm')
plt.figure(figsize = (14,8))
plt.title("AUC-Wilcoxon - AMG_986", fontsize = 24, weight = 'semibold')
heatmap_args = {'linewidths': 0.25, 'linecolor': '0.5', 'clip_on': False, 'square': True, 'cbar_ax_bbox': [0.80, 0.35, 0.04, 0.3]}
sign_plot(pc, **heatmap_args)
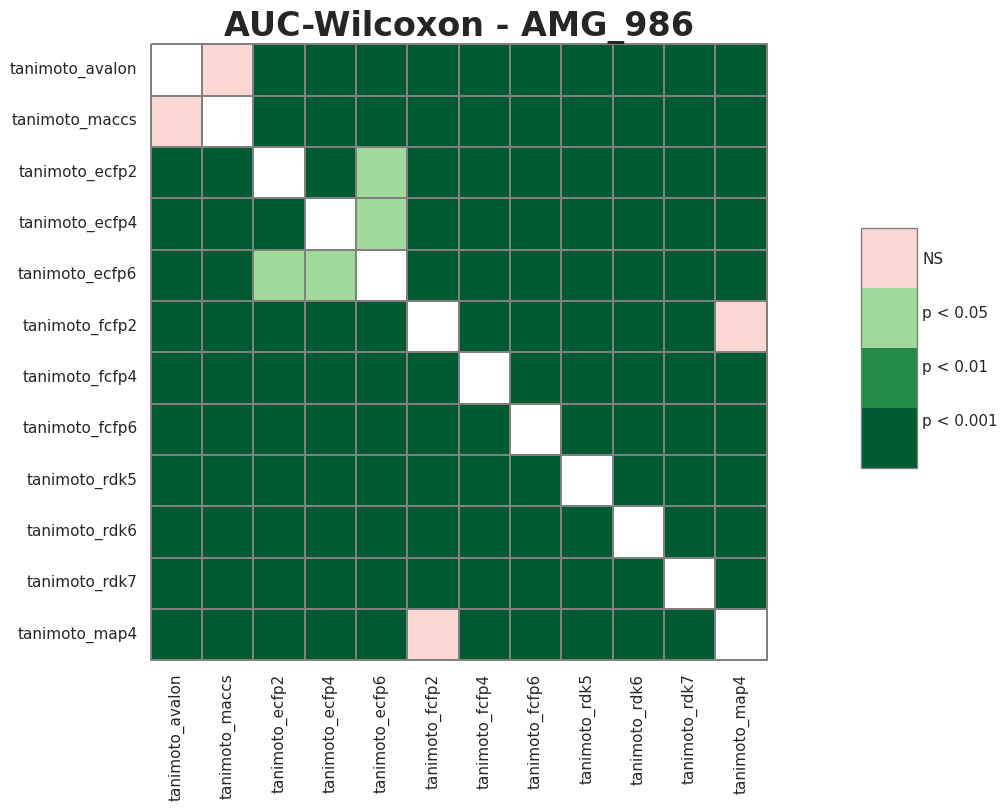
Contributing
Please visit the Mol2DSimi repository. Pull requests are welcome. For major changes, please open an issue first to discuss what you would like to change. Please make sure to update tests as appropriate.
